I’m pleased to report that Paul’s paper on Hyde-CNN has been accepted for publication in Molecular Ecology Resources, and the dadi.CUDA paper is in press at Molecular Biology and Evolution.
We also just posted a major update of our joint DFE preprint. Postdoc Xin Huang did a great job leading this revision. The main content difference is that new large-scale simulations with background selection demonstrate that such linked selection does not bias our inference. Xin also carefully replicated the other results in the paper, consolidating work that had been done by multiple undergraduates over the years to ensure correctness and consistency.
Paul Blischak, a Postdoctoral Fellow working with our group and Mike Barker’s group, just started as Data Scientist at Bayer Crop Science. This is a great position for Paul, combining interesting applied science with the ability to work remotely from anywhere.
We welcome new postdoc Jennifer James to our group. Jenny is coming from Joanna Masel’s group, where she worked on fundamental questions of protein evolution. In our group she’ll be working on applications of joint DFE inference, a callback to her PhD studies with Adam Eyre-Walker.
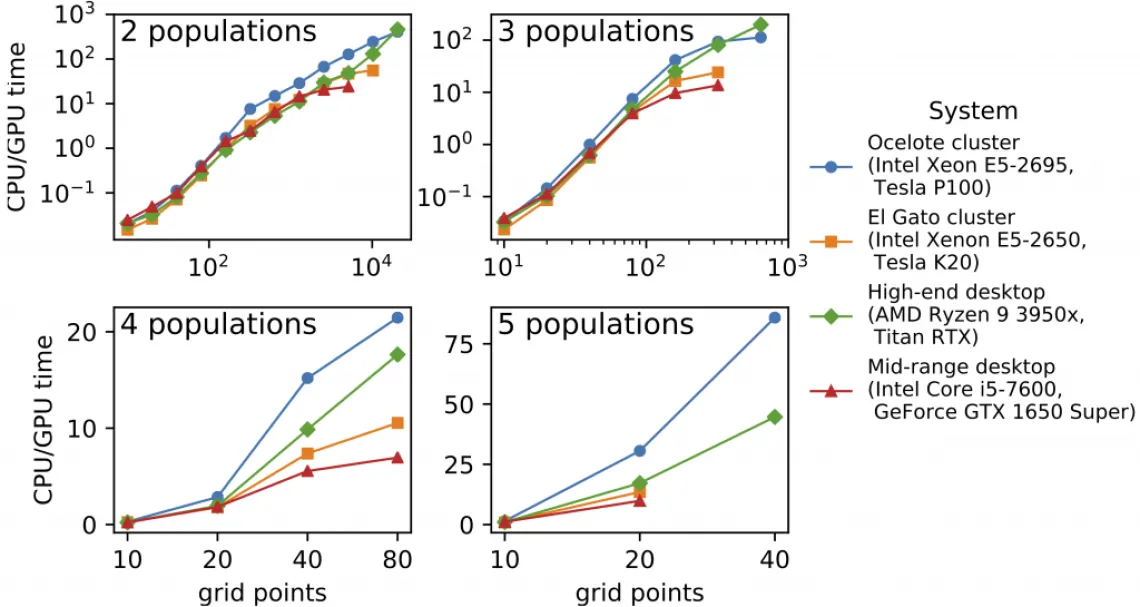
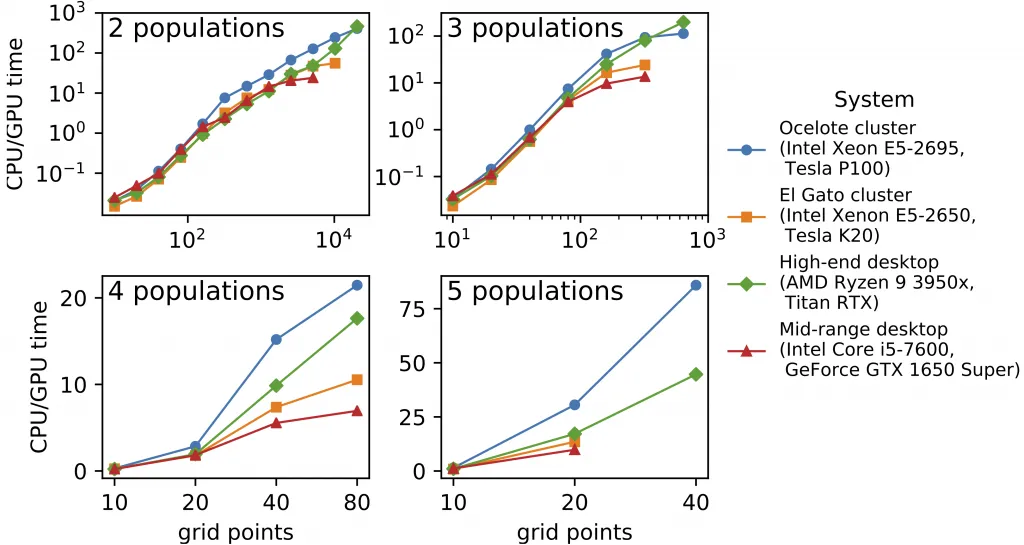
We’ve long known that the core numerical algorithm in dadi is in principle amenable to acceleration through GPU computing. We have now implemented GPU computing in dadi, and the results are spectacular. We see speed ups of over 100x comparing GPUs and CPUs on the same systems. And enabling this speed requires only a single user command. The new feature is described in our bioRxiv preprint, and it is available in dadi 2.1.0.